Personal Inspiration and Project Overview
My journey into this biomedical research began with a personal experiment in supplementation—specifically, with alpha‑lipoic acid. Initially, I was drawn to its reputed benefits, but as I delved deeper into the literature, I discovered astonishing applications. For example, studies have shown that alpha‑lipoic acid can help mitigate diabetic neuropathy and even repair nerve damage in patients. Later, I found another research paper about its benefits in multiple sclerosis, another autoimmune condition, but with fundamentally different mechanisms. These findings sparked a series of questions for me: Could there be deeper, shared biological pathways behind these effects, or are the benefits merely coincidental? With personal and close-to-home experiences of these conditions, I felt compelled to explore the interconnected nature of biological processes and therapeutic responses.
At this time in the AI public conversation sphere, there were coming out more and more articles and research papers about using multiagent systems based on LLMs (Large Language Models) called co-scientists with very promising results in the scientific research area [google research article]. As I already have my hands and experience on this topic because my AI startup generates mobile applications in such multiagentic systems, I could leverage my knowledge by applying it to this domain. So, I decided to start a small project around this topic to learn something. Here is a GitHub repo: AI Bio-researchers
Understanding Multiagent Systems and the Current AI Landscape
Much of the current AI hype revolves around benchmark performances and impressive single-shot abilities of frontier models, often fueling expectations for an eventual artificial general intelligence (AGI). However, this excitement frequently leads to disappointment and criticism when such models fail to produce accurate results consistently. Large Language Models (LLMs) sometimes make mistakes, much like humans do. Yet, humans often overcome initial errors by reflecting and iterating, a capability increasingly mimicked by LLMs when deployed in carefully structured multiagent systems.
Recent research has demonstrated that designing multiple AI agents to collaboratively reason, critique, and refine outputs through defined interaction topologies results in significantly improved robustness and accuracy. Additionally, there’s a misconception that LLMs operate mysteriously or magically, and as they are trained on a human stream of consciousness, they often make such an illusion. In reality, these models are advanced mathematical functions trained to approximate desired outputs given specific numerical inputs, albeit on unprecedented scales.
I believe that by clearly defining the desired outcomes and employing rigorous validation methods, we can significantly enhance the effectiveness and reliability of multiagent LLM systems. Recent advancements, such as DeepSeek’s GRPO algorithm—which uses coding and mathematical tasks (that in nature are verifiable) for reinforcement learning to provide grounded, measurable feedback—illustrate the immense potential of this approach.
In my experimental solution, I chose LangGraph for its exceptional flexibility in controlling agent interactions through graph-based topologies. Its capability to maintain and iteratively update a shared state among agents makes it uniquely suitable for complex research workflows.
Below is a graphical illustration of the multiagent AI bio-researcher topology I have designed:
Graph Workflow Overview:
- Starting Point: The workflow begins with initializing a graph containing basic research details encapsulated in a Research object, including the title and description of the research objectives.
- Knowledge Base Creation Subgraph: Agents reflect on the research goals to determine what research papers will be useful. After downloading them from arXiv, they extract knowledge from papers and synthesize it into a comprehensive Knowledge Graph (KG). I still consider adding other knowledge sources like bioRxiv, PubMed, or general bioscientific books. But this will depend on experiments and how the new knowledge might influence graph construction and retrieval results.
- Research Knowledge Preparation Subgraph (Upcoming): Here, our agents need to explore all available knowledge space in the research topic field. The fact that they have available sources of good-quality biological knowledge (like previously constructed KG) in the knowledge preparation and hypothesis generation phases is very helpful. LLMs are known to hallucinate. While working with agents, we can use this verified, scientific data as some sort of skeleton that they operate on. In the space of ideas and concepts, this data makes them much more likely to operate on things grounded in the real world. They will be less likely to hallucinate, and we will still have the benefit of their creativity, with which they can fill up the gaps between the data or direct it to novel concepts with sophisticated and intelligent thought patterns. Also, we harness the frameworks of intelligent thinking that LLMs learned from vast amounts of data, and additionally, they operate on a huge dimensional space and have somewhere in their parameters some hidden connections between concepts and knowledge in the real world. This balance between operating on real-world grounded scientific facts and this creativity that can generate new ideas and lead to new breakthroughs is crucial. Finding the sweetspot between them, and knowing in what parts need more of this ingredients will determine the success of the project.To ground them in reality, besides KG, I plan to implement more tools. In LangGraph, you can just write a function and use it as a tool. Few examples:
KEGG: a database from which they can query facts about many pathways, interactions, and other.
INDRA: Integrated Network and Dynamical Reasoning Assembler – INDRA draws on natural language processing systems and structured databases to collect mechanistic and causal assertions, represents them in a standardized form (INDRA Statements), and assembles them into various modeling formalisms including causal graphs and dynamical models.
TxGemma: a LLM based on Gemma 2 model, finetuned on therapeutic development knowledge, can predict potential pitfalls (toxicity, trial failure modes). The Reflection agent can incorporate this feedback to weigh translational feasibility. This is very interesting from the point of view that it’s very powerful model that has quite advanced reasoning capabilities but also a lot of biological knowledge.So, agents in this graph have a main planner of the knowledge space traversal. It’s a knowledge space manager. It delegates the research gestions to the specific experts that iterativly reason and use tools to pull knowledge and at the end returns a structured knowledge to the aggregator, that gives it back to the manger who can decide if more knowledge can be discovered or have some gaps that should be filled, or we already have every details and know where the current boundaries of the knowledge are. If so, the subgraph execution finishes, because it is the best place to start generating new research hypotheses.
- Research Hypothesis Generation Subgraph (Upcoming): In this final stage, agents iteratively propose, critique, refine, and validate potential scientific hypotheses, culminating in a comprehensive research report complete with detailed hypotheses and recommended methodologies for empirical validation. In here they can also use the same set of tools when they reason about generated research hypotheses and can cross validate some statements with the tools from which they can acquire data that are grounding AI agents to the real world.
For readers interested in the technical implementation, my project’s detailed graph construction is available on GitHub: research_graph.py.
Enhancing Factuality in LLMs with Graph-Based RAG for Biomedical Research
Ensuring the factual accuracy of large language models (LLMs) remains a significant challenge in today’s GenAI landscape. One of the key approaches to mitigate this issue is Retrieval-Augmented Generation (RAG).
Vanilla RAG: Benefits and Limitations
The vanilla version of RAG works by retrieving chunks of text from a vast corpus and incorporating them into the LLM’s prompt. This technique offers some clear benefits:
- Improved Grounding: By bringing in external text, the model can base its responses on actual data, reducing the likelihood of fabrications.
However, vanilla RAG comes with notable disadvantages:
- Fragmented Context: Chunking text can break key contextual links between related ideas.
- Flat Data Representation: Information is stored in a simplified format, missing complex, layered relationships.
- Limited Context Awareness: Isolated segments prevent the model from grasping full conceptual interdependencies.
In response to these limitations, a host of new RAG techniques are emerging to improve answer quality and maintain contextual integrity.
Graph-Based RAG: Enter LightRAG
To overcome the pitfalls of traditional RAG, my solution employs a graph-based RAG variant, particularly a library called LightRAG. Currently, this approach offers several advantages over other, even more advanced RAG methods:
- Preservation of Context: Rather than operating on disjointed text chunks, graph-based RAG uses a knowledge graph to maintain the full context of the information. This is especially important in biomedical research, where relationships among concepts are intricate and multifaceted.
- Advanced Vector Search: LightRAG leverages sophisticated vector search mechanisms to enhance retrieval speed and overall efficiency.
- Reduced Computational Overhead: Its architecture minimizes the need to rebuild data structures frequently when new data is added, directly reducing operational costs by limiting API calls to LLMs. This is a huge advantage in comparison to Microsoft’s popular library called GraphRAG.
- Intuitive Data Pipeline: The system seamlessly integrates incoming data while automatically de-duplicating entries, ensuring the knowledge graph remains clean and consistent.
This combination of speed, cost-effectiveness, and intelligent data management makes LightRAG an outstanding choice for scalable and robust retrieval-augmented generation applications. For more details, please refer to the LightRAG paper and the LightRAG GitHub Repository.
Why Graph Representation Suits Biology
Biological systems are naturally complex and interconnected. Consider the following characteristics of biological data:
- Hierarchical and Networked: Biological concepts often exist as pathways, circuits, and hierarchical systems—ranging from metabolic pathways and gene regulatory networks to protein interaction maps. All these elements are interrelated, making graph representations an ideal fit.
- Dynamic and Evolving Data: Historically, constructing these graphs required manual annotation by experts—a costly and time-consuming process. With the help of AI, we can now automate this process, continuously updating the knowledge base as new research data becomes available. This automated, real-time updating is crucial in a field where new findings emerge constantly.
Graph construction and knowledge retrieval algorithms
Extraction:
LightRAG begins by breaking the input text into smaller, manageable chunks. Each chunk is transformed into a dense vector embedding that captures its semantic meaning. An LLM is then called to analyze these chunks and extract key entities and relationships, building a structured representation of the content. The extracted data is stored in an indexed format that supports efficient lookup, and built-in de-duplication ensures that redundant or overlapping information is merged.
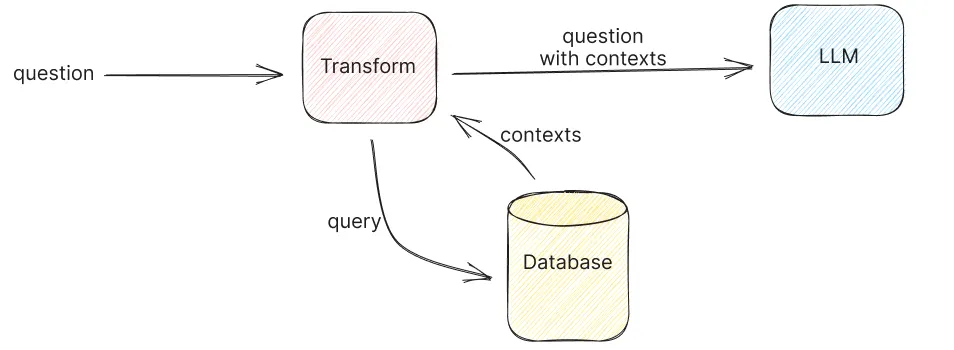
Retrieval:
When a query is made, LightRAG first splits it into essential keywords to identify both local details and broader, global context. It then leverages vector search to compare the query’s embedding against the stored embeddings, quickly pinpointing the most relevant chunks. Finally, a call to an LLM synthesizes the retrieved information into a coherent response that directly addresses the query.
entities extraction:
retrieval:
Experimenting with Knowledge Graph Creation
In developing the knowledge graph for our biomedical research, I encountered several challenges and learned valuable lessons:
- Initial Vanilla Run:
The first attempt used generic prompts designed to extract entities like person, date, place, and action. This approach performed very poorly for biological data, as it failed to capture domain-specific entities and relationships. - Defining Custom Entity Classes:
The library allowed the definition of custom entity classes, which improved results to some extent. However, the framework did not permit manual updates to the entity extraction prompts—such updates could only be applied during the extraction phase, limiting flexibility. Defining custom entity classes only switched the injected into the prompt list of entities that LLM is looking for during the extraction phase. - Modifying the Code Base:
To overcome these limitations, I forked the library from GitHub and edited the code directly. This modification provided the flexibility to manually update prompts, allowing for more biology-centric ontology and prompt injection with biology research–centric examples to significantly enhance the quality of entity extraction.
[You can review the original prompts and the modified prompts on GitHub.] - Integrating Additional Models:
The original library supported only the GPT-4o model family, which proved cost-effective and adequate for the extraction phase. However, for retrieval, I found that the “o” series thinking models (such as o3-mini) delivered better results when paired with customized retrieval prompts.- In one test run, processing approximately 600 research papers cost around $6 using the GPT-4o-mini model combined with OpenAI embeddings.
- Although I did not conduct a large-scale test for the retrieval phase, the API usage remained minimal, keeping costs negligible despite the higher expense of the “o” series models.
This comprehensive approach to constructing a knowledge graph—optimized for the intricate requirements of biological data—demonstrates how modern RAG techniques, when tailored appropriately, can significantly enhance the factual accuracy and relevance of LLM outputs in biomedical research.
Evaluation of the Multiagent System
Building and optimizing a multiagent system is a labor-intensive process that requires numerous manual prompt modifications. In my experience, achieving a quality knowledge graph is a painstaking and somewhat subjective endeavor. Early tests might have seemed promising, but the knowledge space covered by the graph is vast, and gaps in data coverage can easily remain undetected during manual evaluations.
The Challenge of Moving Parts
Each agent in the graph comes with its own set of critical parameters:
- Prompts and Wording:
The phrasing of prompts is crucial. For instance, using polite terms like “please” or “consider” might lead the agent to overlook instructions, whereas more forceful, unambiguous language can compel the agent to act as intended. It is a known and funny fact that if you tell your model that on his response depends your life, it produces better answers… - Data Formatting and Structure:
How raw data is structured—whether through templates, examples, or specific formatting rules—directly impacts the quality of the responses of the LLM. - Model and Parameter Selection:
Each choice of model (gpt-4o, o1-mini, llama, etc), temperature settings, and model parameters tuning can drastically affect outcomes. Small tweaks in one part of the system may yield improvements locally yet trigger suboptimal performance further down the workflow.
This interplay of subtle, cascading effects throughout the system makes it challenging to identify the optimal configuration. Drawing from my own startup experience, where I built multiagent graphs to generate iOS applications that conformed strictly to business requirements and UX/UI designs, I can attest to the frustration of making adjustments that seem promising initially but ultimately lead to poorer results in later stages.
Towards Automated Optimization
Fortunately, the research community is actively addressing these challenges with innovative frameworks designed to reduce the manual overhead of prompt engineering and system configuration. Two notable examples from Stanford are DSPy and TextGrad.
DSPy
- Declarative Task Definition:
Allows you to specify tasks and success metrics in a clear, declarative manner. - Automated Prompt Optimization:
Eliminates manual prompt tweaking by continuously refining prompts using in-context learning and evaluation feedback, leading to more reliable and unbiased model behavior. - ML like mathematical Approach:
Frames prompt engineering as a mathematical optimization problem, thereby reducing human bias and leveraging statistical techniques for improved performance. - Highly Configurable Modules:
Offers flexibility to adjust many elements of the prompting process to suit diverse tasks. - Custom Evaluations:
Lets you define your own evaluation metrics so that the optimization process aligns perfectly with your specific goals. - Optimizing Complete Agent Topologies:
Supports end-to-end optimization of entire pipelines of agents, enabling complex, scalable AI applications.
TextGrad
- Computational Graph Modeling:
Frames the interactions of agents and tools as a computational graph, offering a holistic and optimizable representation of the system. - Dynamic, PyTorch-Like Architecture:
Provides flexibility and an intuitive framework for managing and experimenting with system components. - Mathematical Optimization:
Utilizes optimization algorithms to automatically tune system parameters, eliminating the need for manual prompt engineering. - Custom Evaluation Methods:
Supports the integration of user-defined evaluation metrics to enable tailored feedback and targeted improvements in system performance.
As a machine learning practitioner, I am thrilled to see these frameworks emerge as they offer promising solutions for the challenges inherent in multiagent systems. Their ability to optimize agent configurations mathematically and provide comprehensive quality monitoring is particularly exciting. Given the complexity of these topics, I plan to dedicate a future article specifically to multiagent system mathematical-based optimization. This will not only help in finding the optimal agent configurations but also ensure that our system maintains high quality as it continually updates with new research data.
Summary
In this article, I introduced a multiagent system designed to enhance biomedical research through robust knowledge graph creation and retrieval-augmented generation. By leveraging LightRAG—a graph-based RAG technique that preserves contextual integrity and minimizes computational overhead—I demonstrated how a tailored AI approach can significantly improve factuality in LLM outputs for complex biological domains. The journey involved extensive manual prompt tuning, revealing both the challenges and nuances of optimizing agent configurations. Promising frameworks like DSPy and TextGrad are paving the way for the automated, mathematically driven optimization of these systems.
Stay tuned for two upcoming articles that will delve deeper into the mathematical optimization of agent configurations and further exploration of research subgraphs, pushing the boundaries of AI-driven biomedical discovery.